Poolseq Can Outperform Indiseq
Pool sequencing can outperform individual sequencing
[2020-09-03]
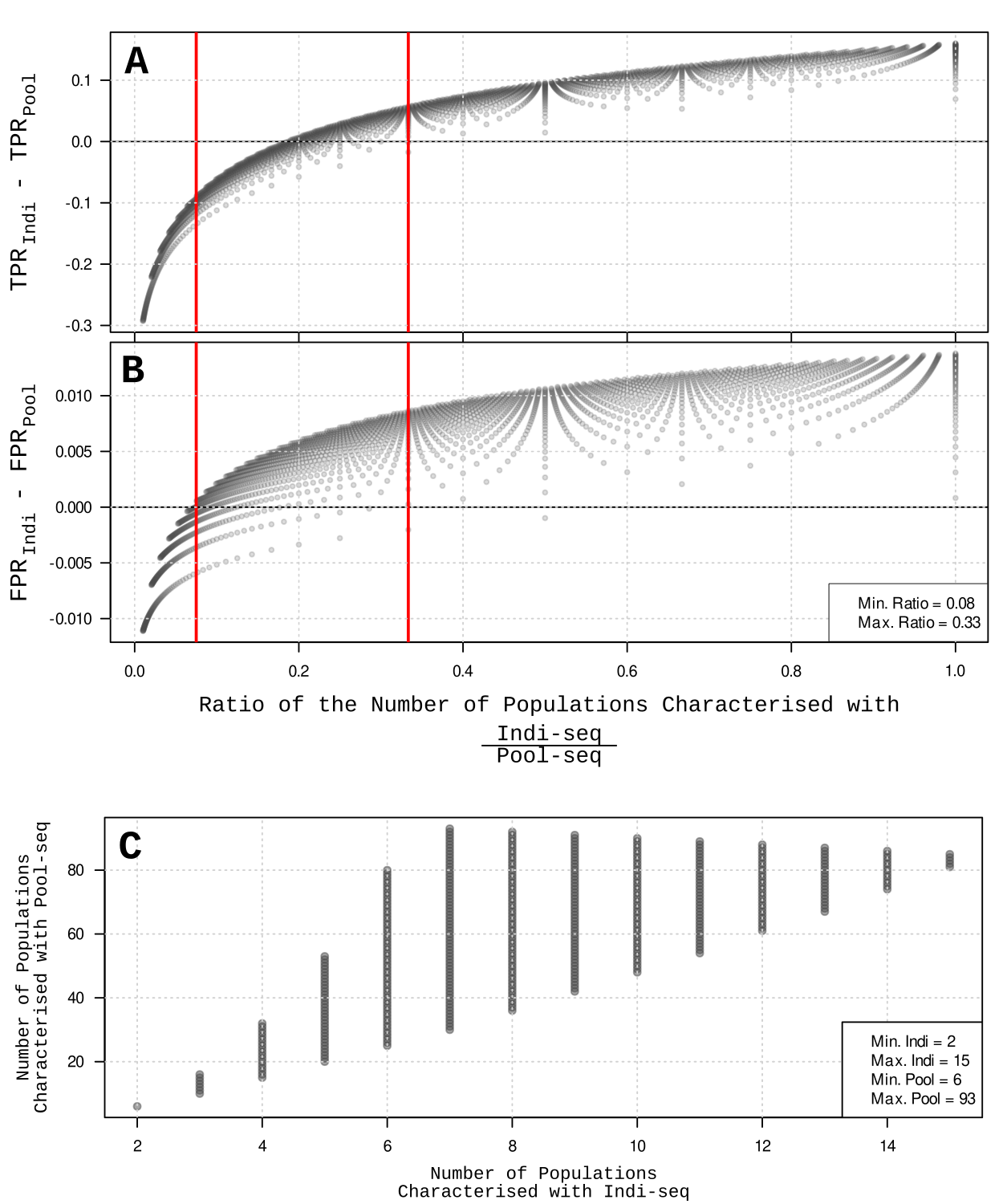
Because pool sequencing (Pool-seq) is always cheaper than individual sequencing (Indi-seq), more populations can be genotyped but at the cost of low resolution (i.e. mutilple individuals are represented by a vector of allele frequencies).
We simulated 405 landscapes under different population genetics parameters and compared GWAS QTL detection accuracies using Pool-seq vs. Indi-seq data as we increase the number of populations sampled. At 1:3 up to 1:12 Indi-seq:Pool-seq population ratios (i.e. for every population genotyped with Indi-seq we can genotype 3 to 12 populations with Pool-seq), GWAS using Pool-seq can achieve higher QTL detection power and lower false positive rate than using Indi-seq.